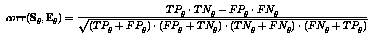When we do multiple tests
at once, as we do in these tables, we have to pay close attention to our
real p-value. For example, if we use p-value limit of 0.05 for three
test, probability of type I error is 0.05+0.05+0.05=0.15 which
is much more that we expected. This is why we have to adjust our test-wise
p-values so we can control our table-wise p-value. Following formulas
allow us to do that:
For independent tests with
tablewise p for n tests: 1-(1-p)^(1/n)
For dependent tests (Bonferoni): p/n
Holm method for j:th test in ascending order of testwise p-values:
p/(n-j+1)
The Holm method (see
for example: Glanz, Slinker A primer of applied regression and analysis
of variance) allows us to increase the discriminative power of multiple
tests.
 Correlation coeffiecient between the set of genes having the binding site
and the set of genes having altered expression. Lets define the notions True
Positive, True Negative, False Positive and False Negative (G=genes, S=genes
w. site, E=genes w. altered expression). Now the correlation coefficient
is
Correlation coeffiecient between the set of genes having the binding site
and the set of genes having altered expression. Lets define the notions True
Positive, True Negative, False Positive and False Negative (G=genes, S=genes
w. site, E=genes w. altered expression). Now the correlation coefficient
is